PCRs
- Polymerase Chain Reaction (PCR) is a technique to amplify specific DNA fragments from their DNA template into multiple copies. Kary Mullis first invented the Polymerase Chain Reaction (PCR) technique in 1983. This technique aims to amplify a DNA fragment from its template to produce a large number of copies.
- The DNA template is obtained from different serological samples present on various substrates, for instance, saliva from a cigarette butt.
Close-up of white DNA helix
- Saliva in the cigarette butt consists of proteins, DNA, lipids, and other enzymes. From handling the sample to isolating the DNA template from the sample, different handling protocols and isolation kits are used to extract high-quality DNA yield. The extracted DNA is then amplified using PCR to generate a higher yield of that DNA template.
- Since the PCR technique is very precise and sensitive, any contamination in the extracted DNA can lead to undesired copies of the expected DNA sample. Hence, care must be taken during the extraction stage to avoid any contamination during the handling and isolation stage.
- PCR technique has a wide range of applications used across various sectors of medical and biological research laboratories. These applications include paternity testing, DNA profiling in forensic investigations, detecting diseases, biotechnology, species identifications, cloning, etc.
Steps in Polymerase Chain Reaction
- Ingredients added in a PCR tube required for PCR reaction are DNA template (target DNA to amplify), two primers (short complementary sequences that attach at the ends of DNA template), Taq polymerase (DNA polymerase to synthesize DNA strands), dNTPs (nucleotides that build to the complementary DNA strand), buffer solution (provides optimum stability to DNA polymerase), and divalent cations (Mg2+ or Mn2+).
- Taq polymerase is a heat-stable enzyme extracted from the bacterium Thermus aquaticus. The role of this DNA polymerase is to attach the dNTPs (nucleotides) to the single-stranded DNA template to create complementary strands to double-stranded DNA fragments.
- Primers for a DNA template can be designed with the help of software that assists in determining suitable primers for that DNA sequence. The two primers are specially designed to complement the targeted DNA fragment to make copies. These primers begin to bind from 3’ to 5’ of the DNA template.
- The primers are designed based on the GC (Guanine and Cytosine) content in the template from the 3’ region. The G and C bases have three H-bonds; therefore, this strong bonding will help the primers to stabilize when binding.
- dNTPs consists of adenine (A), guanine (G), thymine (T), and cytosine (C). These nucleotides continue to bind to the primary single-stranded DNA after primer annealing. Additionally, ddNTPs (dideoxynucleotide triphosphates) are added for chain termination for DNA Sanger sequencing.
- When a DNA polymerase anneals a ddNTP at random, the elongation step ceases. This method is utilised for DNA barcoding, majorly used in species identification.
- Divalent cations, especially Mg2+ from Magnesium Chloride (MgCl2) required to improve the enzymatic activity of DNA polymerase to increase the PCR’s amplification.
Stepwise procedure of PCR’s process
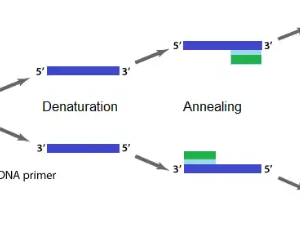
- Denaturation
Denaturation is the first step involving two strands of DNA template being separated or denatured at a high temperature (94-98°C) for 20-30 seconds. This causes the DNA melting to yield a single-stranded DNA template.
- Annealing
Annealing is the process of attaching primers complementary to the single-stranded DNA template at low temperatures (50-70°C) for 20-40 seconds.
- Elongation
Finally, a new DNA strand complementary to the single-stranded DNA template is produced in a 5’ to 3’ direction. The elongation step is also called an extension; upon increasing the temperature (75-80°C), with the help of heat-stable Taq polymerase (a DNA polymerase), it synthesises new DNA strands using dNTPs (deoxynucleoside triphosphates). dNTPs are the building blocks from which DNA polymerase synthesizes new DNA strands.
- Final elongation
This is the last step of PCR’s, where the remaining single-stranded DNA is fully extended for 5-15 minutes at 70-74°C.
- Final hold
This step is optional, where the PCR’s product is kept at 4-15°C for an infinite period as short-term storage of the DNA template.
Close-up of Transferring pink liquid into tubes,
- The PCR’s cycle is repeated 25-35 times to produce billions of DNA copies. After the PCRs process, the amplified DNA template (called an amplimer or amplicon), the concentration and quality of the DNA yield are determined via the gel electrophoresis method. The DNA size is identified against the standard size ladder of known DNA fragment size is run alongside the DNA yield in gel electrophoresis.
- Sometimes, if the primers’ concentration is high, these primers would self-anneal forming primer dimer bands affecting the DNA sequencing. Therefore, a PCR’s clean-up is performed using ExoSAP (Exonuclease I and Shrimp Alkaline Phosphatase) to hydrolyse excess nucleotides and primers.
Types of PCR’s
- There are more than 20 types of PCRs; Real-time PCRs, Multiplex PCRs, Hot Start PCRs, and Reverse Transcriptase PCRs (RT-PCR), to name a few. Real-time PCR is also known as Quantitative PCR. Multiplex PCR is frequently used in molecular biology to amplify multiple targets of the DNA in a single PCR run.
- The amplified DNA is directly proportional to the concentration of DNA in the reaction, hence providing real-time detection of the analysed PCR products. Hot Start PCRs is a new technique aimed at reducing the number of unwanted PCRs products that occurred due to primer-dimer (two primers bind with
- each other to create complementary strands) formation at room temperatures. Reverse Transcriptase PCRs is carried out by converting RNA into complementary DNA, which PCRs then amplifies.
internal links:
What is Genetically Modified organisms vs Transgenic Organisms-Erakina
A novel pathogenic bacterium (Lactobacillus psittaci)-Erakina
28/04/2022